|
| User Guide |
All user interfaces include headers and footers to provide sort and search functions, respectively.
|
|
 Header
Header
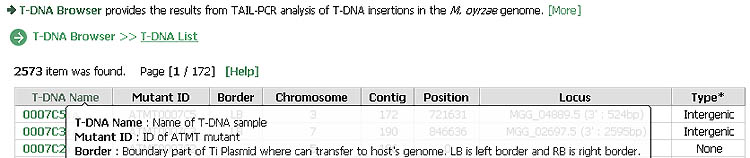
Some field names in a header have links to provide sort function. A first click sorts search results in ascending order while a second click organizes the results in descending order. A third click cancels the sorting. "More" links to a detailed description of each menu and "Help" displays brief information on each field.
|
|
 Footer
Footer
A ˇ°Searchˇ± function is provided in a footer region. For a search, users can select a field and enter a part of description or ID. Users can also choose the number of items to be displayed in one page.

|
The following is a list of all menus included in ATMT database.
|
|
 ATMT Search provides the interface for ATMT mutant searches. A search can be done by a mutant ID or a combination of phenotypic data. A click on each mutant ID accesses tabularized data from high-throughput screening
ATMT Search provides the interface for ATMT mutant searches. A search can be done by a mutant ID or a combination of phenotypic data. A click on each mutant ID accesses tabularized data from high-throughput screening
|
|
 InDepth Browser provides a list of the 559 InDepth mutants with their original mutant IDs and brief descriptions. A click on each InDepth mutant links to its detailed information, such as experimental data from 7 phenotypic analyses and T-DNA-tagged locations (TTLs).
InDepth Browser provides a list of the 559 InDepth mutants with their original mutant IDs and brief descriptions. A click on each InDepth mutant links to its detailed information, such as experimental data from 7 phenotypic analyses and T-DNA-tagged locations (TTLs).
|
|
 T-DNA Browser provides the results from TAIL-PCR analysis of T-DNA insertions. Each sequence ID is linked to detailed information on the T-DNA tag, including its insertional position, locus/loci of affected gene(s) and a diagram. DNA and protein sequences of the affected gene can also be retrieved from our in-house database. Original sequence information was imported from the Magnaporthe grisea database at Broad Institute.
T-DNA Browser provides the results from TAIL-PCR analysis of T-DNA insertions. Each sequence ID is linked to detailed information on the T-DNA tag, including its insertional position, locus/loci of affected gene(s) and a diagram. DNA and protein sequences of the affected gene can also be retrieved from our in-house database. Original sequence information was imported from the Magnaporthe grisea database at Broad Institute.
|
|
 Chromosome Viewer illustrates the distribution of TTLs on each chromosomes of M. oryzae. M. oryzae chromosomal information for genetic and optical mapping of TTLs was provided by genome database release 5 from Broad Institute. Each red line indicates one TTL insertion and numbers at bottom indicate the total numbers of TTLs on each chromosomes. UA indicates a collection of unassigned TTLs.
Chromosome Viewer illustrates the distribution of TTLs on each chromosomes of M. oryzae. M. oryzae chromosomal information for genetic and optical mapping of TTLs was provided by genome database release 5 from Broad Institute. Each red line indicates one TTL insertion and numbers at bottom indicate the total numbers of TTLs on each chromosomes. UA indicates a collection of unassigned TTLs.
|
|
 Density Viewer displays the density of TTLs in every 100kb chromosomal region and offers graphical comparison of TTL densities among chromosomes.
Density Viewer displays the density of TTLs in every 100kb chromosomal region and offers graphical comparison of TTL densities among chromosomes.
|
| Glossary |
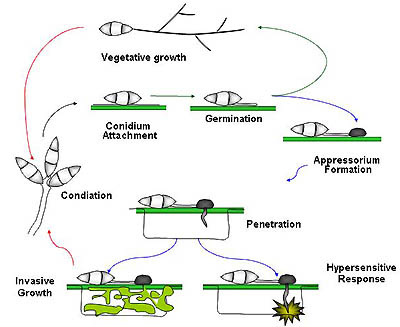
 Disease cycle of Magnaporthe oryzae
Disease cycle of Magnaporthe oryzae
|
|
 ATMT (Agrobacterium tumefaciens-meidated transformation) is an efficient tool for insertional mutagenesis used in many model organisms. In addition to high rates of single copy insertion of T-DNA (Rho et al., 2002), its big advantage over traditional chemical or radiation mutagenesis procedures is that the mutated genes are tagged by T-DNA and can subsequently be cloned using the T-DNA.
ATMT (Agrobacterium tumefaciens-meidated transformation) is an efficient tool for insertional mutagenesis used in many model organisms. In addition to high rates of single copy insertion of T-DNA (Rho et al., 2002), its big advantage over traditional chemical or radiation mutagenesis procedures is that the mutated genes are tagged by T-DNA and can subsequently be cloned using the T-DNA.
|
|
 T-DNAmeans the transferred DNA of the Ti plasmid vector of Agrobacterium tumefacines. The genes that are located in the T-DNA region contain the necessary structural features for expression in a transformed cell. The key elements of the T-DNA are the 25-base pair imperfect repeats present at the boundaries of the T-DNA (left border (LB) and right border (RB)).
T-DNAmeans the transferred DNA of the Ti plasmid vector of Agrobacterium tumefacines. The genes that are located in the T-DNA region contain the necessary structural features for expression in a transformed cell. The key elements of the T-DNA are the 25-base pair imperfect repeats present at the boundaries of the T-DNA (left border (LB) and right border (RB)).
|
|
 TAIL-PCR (Thermal Asymmetric Interlaced-PCR) is a powerful method to recover unknown DNA fragments adjacent to known sequences of T-DNA or transposon insertions (Liu et al., 1995; Smith et al., 1996). It uses nested, insertion-specific primers together with arbitrary degenerate (AD) primers having lower annealing temperatures. Relative amplification efficiencies of specific products and non-specific products are thermally controlled through alternating cycles of high and low annealing temperature. In the primary reaction, one low-stringency PCR cycle is conducted to create one or more annealing sites for an AD primer in the targeted sequence. A specific product is then preferentially amplified over nonspecific ones by interspersion of two high-stringency PCR cycles with one reduced-stringency PCR cycle. The nested PCR amplifications help to achieve higher specificity (Liu and Huang, 1998).
TAIL-PCR (Thermal Asymmetric Interlaced-PCR) is a powerful method to recover unknown DNA fragments adjacent to known sequences of T-DNA or transposon insertions (Liu et al., 1995; Smith et al., 1996). It uses nested, insertion-specific primers together with arbitrary degenerate (AD) primers having lower annealing temperatures. Relative amplification efficiencies of specific products and non-specific products are thermally controlled through alternating cycles of high and low annealing temperature. In the primary reaction, one low-stringency PCR cycle is conducted to create one or more annealing sites for an AD primer in the targeted sequence. A specific product is then preferentially amplified over nonspecific ones by interspersion of two high-stringency PCR cycles with one reduced-stringency PCR cycle. The nested PCR amplifications help to achieve higher specificity (Liu and Huang, 1998).
|
|
 TTL (T-DNA-Tagged Location) is defined as a DNA region containing one or more T-DNA insertions.
TTL (T-DNA-Tagged Location) is defined as a DNA region containing one or more T-DNA insertions.
|
|
 InDepth is a subset of ATMT mutants selected via high-throughput screening based on their pathogenicity defects.
InDepth is a subset of ATMT mutants selected via high-throughput screening based on their pathogenicity defects.
|
|
|
|
|
|